A Least Squares Approach
The essentials
Choose w to get the min Loss function.
we can change to fit our needs, but here we use the mostly used square root sum function.
Turning point
Goal: to choose the minimum in a mathmatical way. How it works: the partial derivatives of should be , and second derivatives should be a positive value to ensure this is a minimum/maxmum
Example
#data.csv
n xn tn xntn x2
1 1896 12.00 22752.0 3.5948*10**6
2 1900 11.00 20900.0 3.6100*10**6
3 1904 11.00 20944.0 3.6252*10**6
4 1906 11.20 21347.2 3.6328*10**6
5 1908 10.80 20606.4 3.6405*10**6
6 1912 10.80 20649.6 3.6557*10**6
7 1920 10.80 20736.0 3.6864*10**6
8 1924 10.60 20394.4 3.7018*10**6
9 1928 10.80 20822.4 3.7172*10**6
10 1932 10.30 19899.6 3.7326*10**6
11 1936 10.30 19940.8 3.7481*10**6
12 1948 10.30 20064.4 3.7947*10**6
13 1952 10.40 20300.8 3.8103*10**6
14 1956 10.50 20538.0 3.8259*10**6
15 1960 10.20 19992.0 3.8416*10**6
16 1964 10.00 19640.0 3.8573*10**6
17 1968 9.95 19581.6 3.8730*10**6
18 1972 10.14 19996.1 3.8888*10**6
19 1976 10.06 19878.6 3.9046*10**6
20 1980 10.25 20295.0 3.9204*10**6
21 1984 9.99 19820.2 3.9363*10**6
22 1988 9.92 19721.0 3.9521*10**6
23 1992 9.96 19840.3 3.9681*10**6
24 1996 9.84 19640.6 3.9840*10**6
25 2000 9.87 19740.0 4.0000*10**6
26 2004 9.85 19739.4 4.0160*10**6
27 2008 9.69 19457.5 4.0321*10**6
# python3
import numpy as np
import pandas as pd
import copy as copy
import matplotlib.pyplot as plt
df = pd.read_csv('data.csv',sep=' ')
mm = copy.deepcopy(df)
for index, row in df.iterrows():
df.loc[index,'x2'] = eval(df.iloc[index]['x2'])
print(int(df.iloc[index]['x2']))
mean = df.mean()
w1 = (mean['xntn']-mean['xn']*mean['tn'])/(mean['x2'] -mean['xn']*mean['xn'])
w0 = mean['tn'] - w1*mean['xn']
xn = mm.loc[:,'xn']
tn = mm.loc[:,'tn']
x = np.linspace(1870,2020,100) # 100 linearly spaced numbers
y = w0 + w1*x
plt.xlabel('Years', fontsize = 30)
plt.ylabel('Time(second)', fontsize = 30)
plt.plot(xn, tn, 'ro')
plt.plot(x,y)
plt.show()
Result:
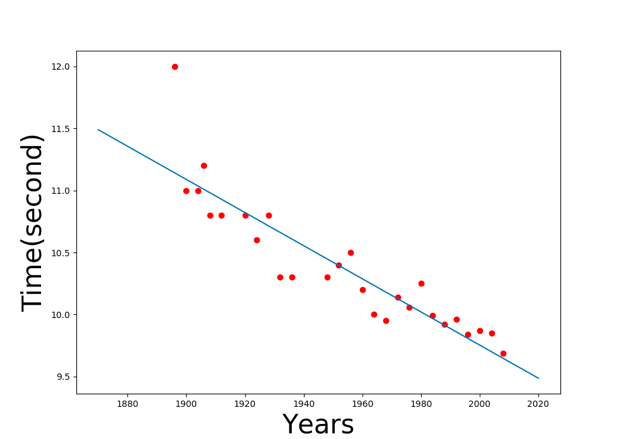 Linear model
Linear model
Vector/ Matrix notation
why?
- To express many variables in one time example:
useful identities:
Squre root
# data is the same as above
import numpy as np
import pandas as pd
import copy as copy
import matplotlib.pyplot as plt
df = pd.read_csv('data.csv',sep=' ')
mm = copy.deepcopy(df)
for index, row in df.iterrows():
df.loc[index,'x2'] = eval(df.iloc[index]['x2'])
#print(int(df.iloc[index]['x2']))
mean = df.mean()
X = np.ones(mm.shape[0]*2).reshape(mm.shape[0],2)
t = np.ones(mm.shape[0]).reshape(mm.shape[0],1)
for index in range(X.shape[0]):
X[index][1] = mm.loc[index, 'xn']
t[index][0] = mm.loc[index, 'tn']
Xt = np.transpose(X)
w = np.dot(np.dot(np.linalg.inv(np.dot(Xt, X)) , Xt),t)
xn = mm.loc[:,'xn']
tn = mm.loc[:,'tn']
x = np.linspace(1870,2020,100)
y = w[0] + w[1]*x
plt.xlabel('Years', fontsize = 30)
plt.ylabel('Time(second)', fontsize = 30)
plt.plot(xn, tn, 'ro')
plt.plot(x,y)
plt.show()
# print(w)
# out: array([[3.64164559e+01],[-1.33308857e-02]])
# the same result with above
# another better method and poly
import pylab as plt
import urllib.request
# You only need this line if you haven't cloned the repo...if you have cloned, you'll already have the data
urllib.request.urlretrieve('https://raw.githubusercontent.com/sdrogers/fcmlcode/master/notebooks/data/olympic100m.txt', 'olympic100m.txt')
import numpy as np
# If you have cloned, make sure this is pointing to the correct file, maybe ../data/olympic100m.txt ?
data = np.loadtxt('olympic100m.txt',delimiter=',')
x = data[:,0][:,None]
t = data[:,1][:,None]
X = np.hstack((np.ones_like(x),x,x**2)) # we just add more to here to change all
X = np.asmatrix(X)
t = t # This is already a vector!
w = (X.T*X).I*X.T*t
testx = np.linspace(1896,2012,100)[:,None]
testX = np.hstack((np.ones_like(testx),testx,testx**2))
testt = testX*w
plt.figure()
plt.plot(x,t,'ro')
plt.plot(testx,testt,'b')
plt.show()
…
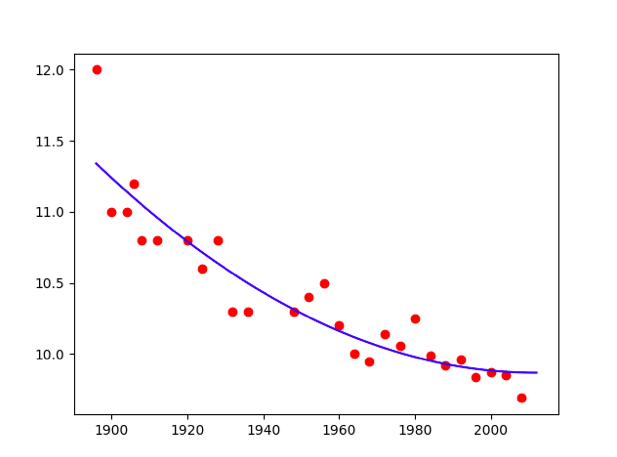 Poly = 2
Poly = 2 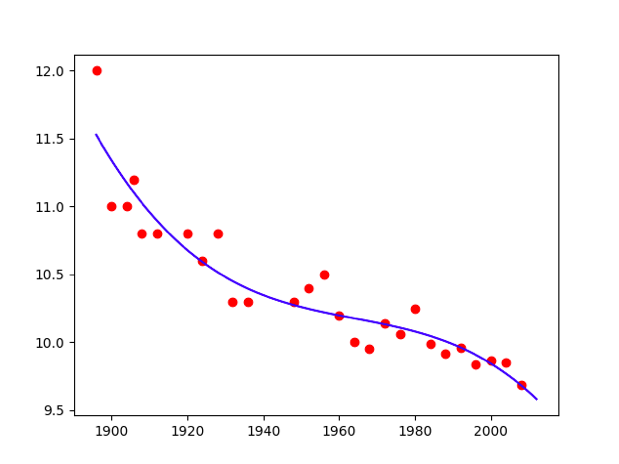 Poly = 3
Poly = 3
we are free to define any set of K functions of X
Like:
import pylab as plt
import urllib.request
# You only need this line if you haven't cloned the repo...if you have cloned, you'll already have the data
urllib.request.urlretrieve('https://raw.githubusercontent.com/sdrogers/fcmlcode/master/notebooks/data/olympic100m.txt', 'olympic100m.txt')
import numpy as np
import math
# If you have cloned, make sure this is pointing to the correct file, maybe ../data/olympic100m.txt ?
data = np.loadtxt('olympic100m.txt',delimiter=',')
x = data[:,0][:,None]
t = data[:,1][:,None]
hx = np.sin((x-2660)/4.3)
X = np.hstack((np.ones_like(x),x,hx)) # we just add more to here to change all
X = np.asmatrix(X)
t = t # This is already a vector!
w = (X.T*X).I*X.T*t
testx = np.linspace(1896,2012,100)[:,None]
testX = np.hstack((np.ones_like(testx),testx,np.sin((testx-2660)/4.3)))
testt = testX*w
plt.figure()
plt.plot(x,t,'ro')
plt.plot(testx,testt,'b')
plt.show()
… 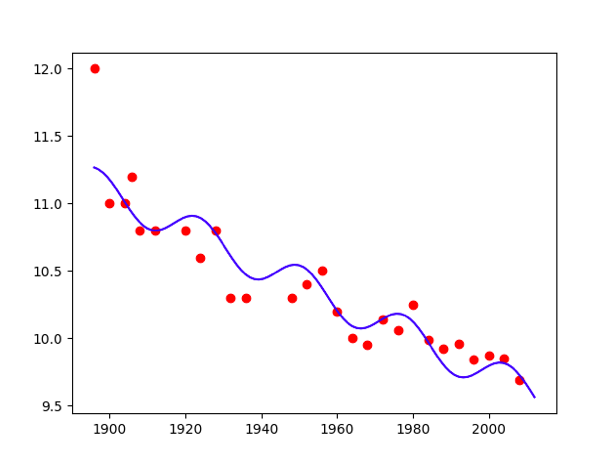 Polynomial model
Polynomial model
Validation data
- Why we need to validate?
- to restrain overfitting
- How we validate data?
- a effecient technique – Cross-validation(K-fold)
- How to prevent from overfitting?
- regularisation- if we don’t want our model to become too complex
- regularisation- if we don’t want our model to become too complex
import pylab as plt
import urllib.request
# You only need this line if you haven't cloned the repo...if you have cloned, you'll already have the data
urllib.request.urlretrieve('https://raw.githubusercontent.com/sdrogers/fcmlcode/master/notebooks/data/olympic100m.txt', 'olympic100m.txt')
import numpy as np
import math
# If you have cloned, make sure this is pointing to the correct file, maybe ../data/olympic100m.txt ?
data = np.loadtxt('olympic100m.txt',delimiter=',')
x = data[:,0][:,None]
t = data[:,1][:,None]
maxorder = 7
x_test = np.linspace(1896,2020,100)[:,None]
X = np.ones_like(x)
X_test = np.ones_like(x_test)
for i in range(1,maxorder+1):
X = np.hstack((X,x**i))
X_test = np.hstack((X_test,x_test**i))
X = np.asmatrix(X)
for lamb in [0,0.01,0.1,1,10,100]:
w = np.linalg.solve(np.dot(X.T,X) + x.size*lamb*np.identity(maxorder+1),np.dot(X.T,t))
A = np.dot(X.T,X)
B = np.dot(X.T,t)
m = A.I*B
#w = np.linalg.solve(A,B)
f_test = np.dot(X_test,w)
plt.figure()
plt.plot(x_test,f_test,'b-',linewidth=2)
plt.plot(x,t,'ro')
title = '<span class="katex"><span class="katex-mathml"><math xmlns="http://www.w3.org/1998/Math/MathML"><semantics><mrow><mi>λ</mi><mo>=</mo></mrow><annotation encoding="application/x-tex">\lambda=</annotation></semantics></math></span><span class="katex-html" aria-hidden="true"><span class="base"><span class="strut" style="height:0.69444em;vertical-align:0em;"></span><span class="mord mathdefault">λ</span><span class="mspace" style="margin-right:0.2777777777777778em;"></span><span class="mrel">=</span></span></span></span>%g'%lamb
plt.title(title)
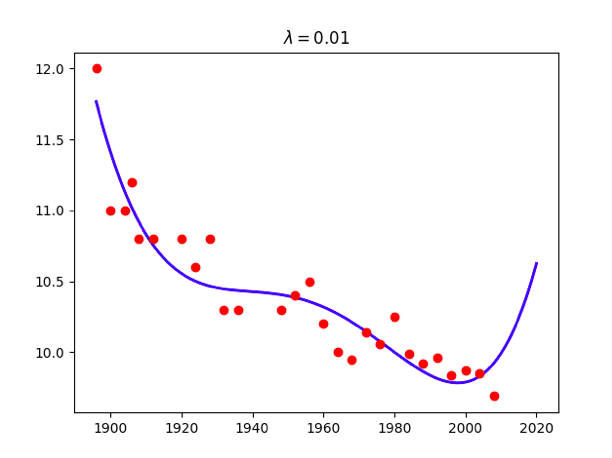
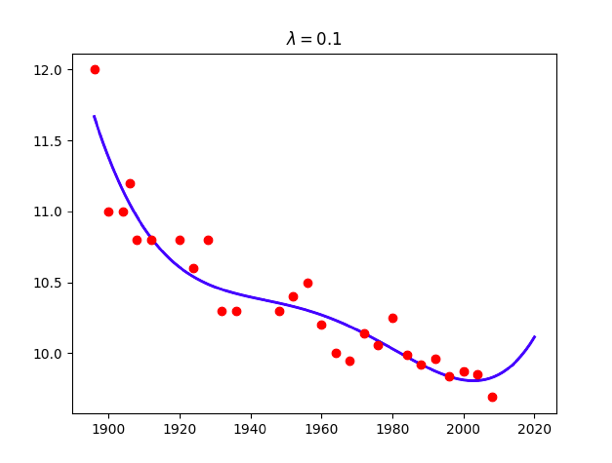
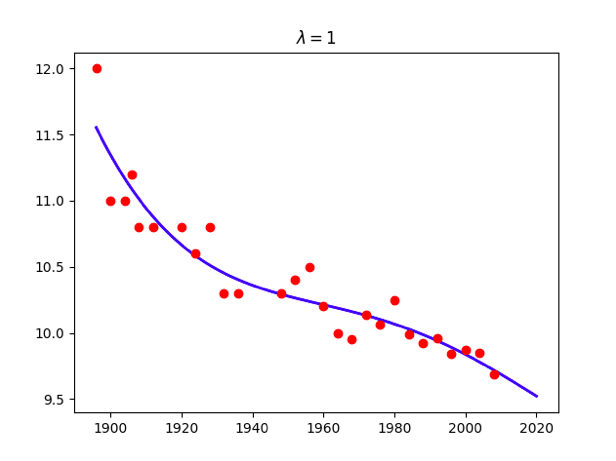
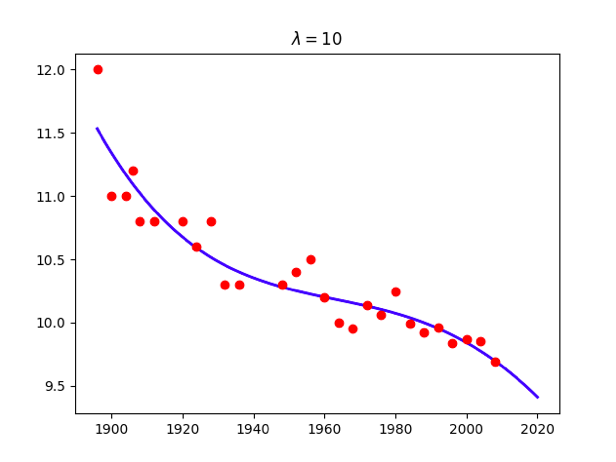
Attention: use np.linalg.solve(A,B) instead of becauese the accuracy problem.
Cross-validation
The loss that we calculate from validation data will be sensitive to the choice of data in our validation set, especially dataset is small. so we need a method to make more efficient use of the data we have K-fold cross-validation, particularly, Leave-One-Out Cross-Validation(LOOCV)
The best model – minimize test loss
import pylab as plt
import urllib.request
# You only need this line if you haven't cloned the repo...if you have cloned, you'll already have the data
urllib.request.urlretrieve('https://raw.githubusercontent.com/sdrogers/fcmlcode/master/notebooks/data/olympic100m.txt', 'olympic100m.txt')
import numpy as np
import math
# If you have cloned, make sure this is pointing to the correct file, maybe ../data/olympic100m.txt ?
data = np.loadtxt('olympic100m.txt',delimiter=',')
x = data[:,0][:,None]
t = data[:,1][:,None]
maxorder = 8
x_train = x[:-3]
x_test = x[-3:]
t_train = t[:-3]
t_test = t[-3:]
X_train = np.ones_like(x_train)
X_test = np.ones_like(x_test)
errorlist = []
errorstdlist = []
for i in range(1,maxorder+1):
X_train = np.hstack((X_train,x_train**i))
X_test = np.hstack((X_test,x_test**i))
w = np.linalg.solve(np.dot(X_train.T,X_train) ,np.dot(X_train.T,t_train))
#w = np.linalg.solve(A,B)
f_test = np.dot(X_test,w)
error_std = np.std(f_test - t_test)
errorstdlist.append(np.std(np.dot(X_train,w) - t_train))
errorlist.append (error_std)
f_test = np.dot(X_test,w)
plt.figure()
plt.plot([1,2,3,4,5,6,7,8],errorlist,'b-',linewidth=2)
plt.plot([1,2,3,4,5,6,7,8],errorstdlist,'y-',linewidth=2)
plt.show()
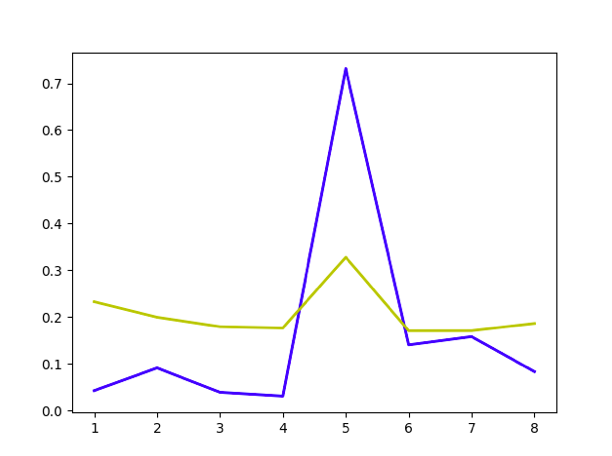 Best model visualization Then we can choose the best model.
Best model visualization Then we can choose the best model.
LOOCV loss and train, test less
LOOCV loss: 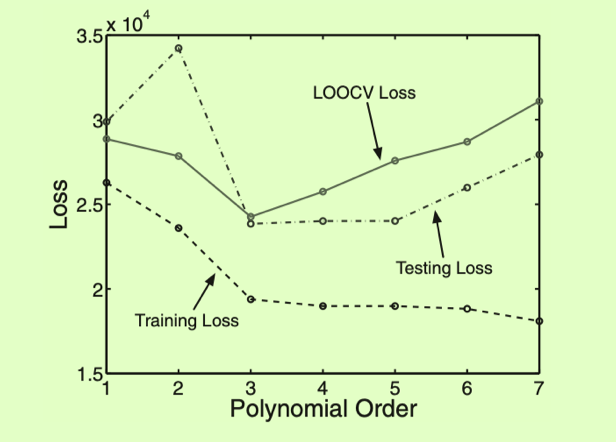
Welcome to share or comment on this post: