2019 September 04 Machine Learning, Math
The Bayesian Approach To Machine Learning
The likelihood
p(t∣w,X,σ2)=N(Xw,σ2IN)
The prior
p(w∣μ0,Σ0)=N(μ0,Σ0) We need to choose μ0 and Σ0
The posterior
p(w∣t,X,σ2)=∝p(t∣w,X,σ2)p(w∣μ0)(2π)N/2∣σ2I∣1/21exp(−21(t−Xw)⊤(σ2I)−1(t−Xw))×(2π)N/2∣Σ0∣1/21exp(−21(w−μ0)⊤Σ0−1(w−μ0))
∝=exp(−2σ21(t−Xw)⊤(t−Xw))×exp(−21(w−μ0)⊤Σ0−1(w−μ0))exp{−21(σ21(t−Xw)⊤(t−Xw)+(w−μ0)⊤Σ0−1(w−μ0))}
p(w∣t,X,σ2)∝exp{−21(−σ22t⊤Xw+σ21w⊤X⊤Xw+w⊤Σ0−1w−2μ0⊤Σ0−1w)}
p(w∣t,X,σ2)=N(μw,Σw)∝exp(−21(w−μw)⊤Σw−1(w−μw))∝exp{−21(w⊤Σw−1w−2μw⊤Σw−1w)}
The terms linear and quadratic in w in equations must be equal, so
⇒ w⊤Σw−1w=σ21w⊤X⊤Xw+w⊤Σ0−1w=w⊤(σ21X⊤X+Σ0−1)w Σw=(σ21X⊤X+Σ0−1)−1
and −2μw⊤Σw−1wμw⊤Σw−1w=−σ22t⊤Xw−2μ0⊤Σ0−1w=σ21t⊤Xw+μ0⊤Σ0−1w μw⊤Σw−1=σ21t⊤X+μ0⊤Σ0−1 μwTΣw−1Σw=(σ21t⊤X+μ0⊤Σ0−1)Σw μw⊤=(σ21t⊤X+μ0⊤Σ0−1)Σw μw=Σw(σ21XTt+Σ0−1μ0)
import numpy as np
import random
import pylab as plt
def gaussian2d(mu,sigma,xvals,yvals):
const = (1.0/(2.0*np.pi))*(1.0/np.sqrt(np.linalg.det(sigma)))
si = np.linalg.inv(sigma)
xv = xvals-mu[0]
yv = yvals-mu[1]
return const * np.exp(-0.5*(xv*xv*si[0][0] + xv*yv*si[1][0] + yv*xv*si[0][1] + yv*yv*si[1][1]))
if __name__ == '__main__':
data = np.loadtxt('olympic100m.txt',delimiter=',')
x = data[:,0][:,None]
t = data[:,1][:,None]
x = x - x[0]
x/= 4.0
t = t # This is already a vector!
inputx = x.reshape(len(x),1)
X = np.array([[1,inputx[0]]])
T = np.array([t[0]])
sig = np.array([[100,0],[0,5]])
myu = np.array([[0],[0]])
xp = np.arange(-30,30,0.1)
yp = np.arange(-10,10,0.1)
Xp,Yp = np.meshgrid(xp,yp)
Z = gaussian2d(myu,sig,Xp,Yp)
#CS = plt.contour(Xp,Yp,Z,20,colors='k')
#plt.xlabel('w_0')
#plt.ylabel('w_1')
sig_sq = 2
plt.figure()
plt.scatter(x,t)
#Y = y.reshape(len(y),1)
count = 0
xpsub = np.arange(9,13,0.02)
ypsub = np.arange(-0.5,0.5,0.02)
Xpsub,Ypsub = np.meshgrid(xp,yp)
for addx, addt in zip(inputx[1:], t[1:]):
#print(X,T)
count += 1
T = np.vstack((T,addt))
X = np.vstack((X,[1,addx]))
#signew = np.linalg.inv((1.0/sig_sq)*np.dot(X.T,X) + np.linalg.inv(sig))
#myunew = np.dot(signew,(1.0/sig_sq)*np.dot(X.T,T) + np.dot(np.linalg.inv(sig),myu))
signew = np.linalg.inv((1.0/sig_sq)*X.T@X + np.linalg.inv(sig))
myunew = signew@((1.0/sig_sq)*X.T@T + np.linalg.inv(sig)@myu)
#print(myunew)
if count%2== 0:
plt.figure()
plt.subplot(1,2,1)
xl = np.linspace(-5,30,100)
xline = xl.reshape(len(xl),1)
xline = np.hstack((np.ones_like(xline),xline))
yline = xline@myunew
plt.plot(xl, yline)
plt.scatter(x,t)
plt.subplot(1,2,2)
zposterior = gaussian2d(myunew, signew, Xpsub, Ypsub)
cs = plt.contour(Xp, Yp,Z,20, colors = 'k')
cs = plt.contour(Xpsub, Ypsub, zposterior,colors='r')
plt.xlim([9,13])
plt.ylim([-0.5,0.5])
plt.title('First' + str(count) + ' points, contours')
plt.show()
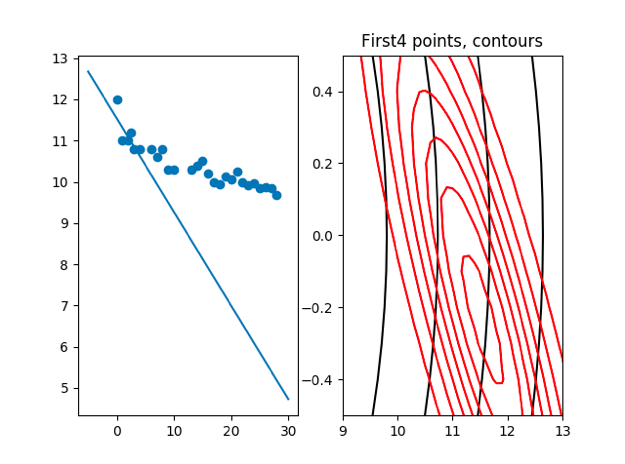
cov:[1.47783251,−0.82101806] [−0.82101806,0.826491521] μ: [[11.57635468] [−0.32019704]]
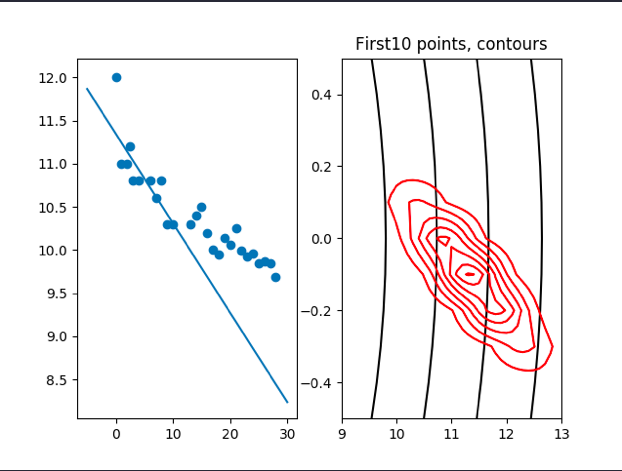
cov:[0.6722373,−0.12130803] [−0.12130803,0.03266264] μ: [[11.32030208] [−0.09280969]]
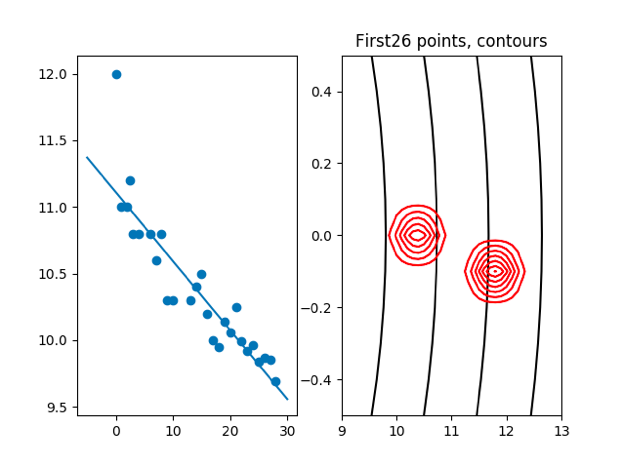
cov: [0.26539973−0.01359028] [−0.01359028,0.00096507]
we can see that the cov is becomming more and more small with the incresing of points, that means we are less affected by the prior μ and cov.
plt.scatter(x,t)
xnew = np.linspace(-5,50,100)
xnew = xnew.reshape(len(xnew),1)
xerror = np.ones_like(xnew)
#for m in range(1,i+1):
xerror = np.hstack((xerror,xnew))
inverse = np.linalg.inv(X.T@X)
newsigma = sig_sq*xerror@inverse@xerror.T
newsigma = np.diag(newsigma)
plt.errorbar(xnew,xerror@myunew,yerr=newsigma)
plt.show()
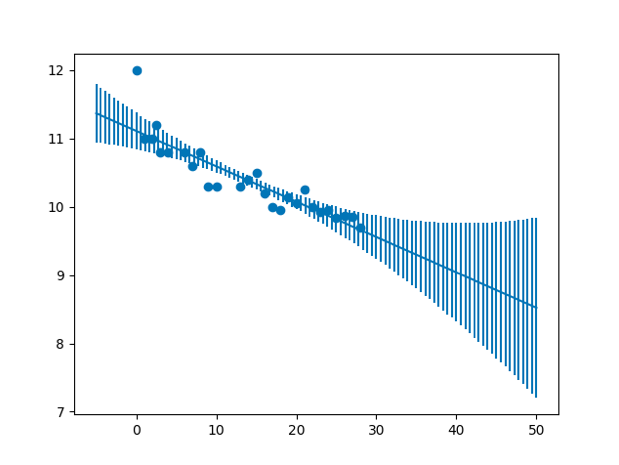 Variance of the model
Variance of the model
Welcome to share or comment on this post:



 Variance of the model
Variance of the model